Synthesis and Crystal Structure of Ethyl 6-(chloromethyl)-4-(3-Chlorophenyl)-2-oxo- 1,2,3,4-Tetrahydropyrimidine-5-Carboxylate
S Bharanidharan, H Saleem1, M Suresh and B Gunasekaran
S Bharanidharan1, H Saleem1, M Suresh2 and B Gunasekaran3*
1Department of Physics, Annamalai University, Annamalainagar-608 002, Tamil Nadu, India
2Department of Chemistry, LN Government Chemistry (Autonomous), Ponneri-601 204, Tamil Nadu, India
3Department of Physics and Nano Technology, SRM University, SRM Nagar, Kattankulathur Campus, Chennai 603 203, Tamil Nadu, India
- *Corresponding Author:
- Gunasekaran B
Department of Physics and Nano Technology, SRM University, SRM Nagar, Kattankulathur Campus, Chennai 603 203, India
Tel: + 91 9962525891
E-mail:gunasekaran.b@ktr.srmuniv.ac.in; bguna_sekaran77@yahoo.co.in
Received Date: September 11, 2015; Accepted Date: October 06, 2015; Published Date: October 25, 2015
Keywords
Single-crystal X-ray study; Tetrahydropyrimidine; Weak interaction; R factor
Introduction
In recent years, the interest in Dihydropyrimidines (DHPMs) has increased rapidly because of the structural resemblance of DHPM with clinically important Hantzsch pyridines [1,2]. The biologically active dihydropyridine molecules contain the substituent 4-phenyl ring positioned above and in the vertical plane of 1, 4-dihydropyridine ring, which itself is a flattened boat conformation [3]. Pyrimidine derivatives comprise a diverse and interesting group of drugs which are extremely important for their biological activities. Dihydropyrimidines and their derivatives have attracted increasing interest owing to their therapeutic and pharmaceutical properties, such as antiviral, antitubercular, [4,5] antimicrobial agent [6-10], antagonists of the human adenosine A2A receptor [11], cyclooxygenase-2 inhibitory activity [12,13], tyrosine kinase inhibitors [14], Antiamoebic activity [15], and cytotoxicity [16,17]. The chemical structure of sulphanilamide provides a most valuable molecular template for the development of agents able to interact with a wide variety of biological activities [18]. The tetrahydropyrimidines is structurally similar to Dihydropyrimidines. Hence, it was thought worthwhile to synthesize new congeners by incorporating chlorophenyl and carboxylate with 1,2,3,4-tetrahydropyrimidinones moieties in a single molecular framework.
Experimental
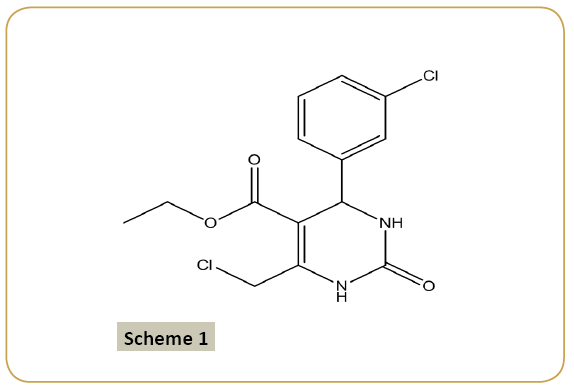
A mixture of ethyl-4-chloroacetoacetate (4.1 mL, 0.025 mol), 3-chlorobenzaldehyde (3.6 gm, 0.025 mol), and urea (4.5 g, 0.075 mol) in ethanol (5 ml) was heated under reflux in the presence of concentrated HCl (1 mL) for 8 h (monitored by TLC). The reaction mixture, after being cooled to room temperature, was poured onto crushed ice and stirred for 5-10 min. The precipitate was washed with sodium bicarbonate solution, filtered, dried and again washed with petroleum ether (40-60%) and dried over in a vacuum. The compound was recrystallized from absolute ethanol with melting point 151-153≡ C and yield 72% (Scheme 1).
X-ray structure determination
Single crystal X-ray diffraction data for the compound at room temperature was collected by Bruker Kappa diffractometer with Mo Kα radiation using ω/2θ scan mode. SMART APEX2 CCD area detector with Mo Kα radiation and ω scan mode was applied to obtain an accurate unit cell parameters and orientation matrix within the least-square fit of several high angle reflections in the ranges 2.3 ≡<θ<19.20 ≡. Cell refinement and data reduction were carried out using SAINT. A total of 4647 reflections were collected, resulting in 1230 independent reflections of which 905 had I>2σ(I). The intensities for Lorentz and polarization effects and absorption corrections were corrected by using SADABS [19]. The structure of compound was solved by direct method procedure as implemented in SHELXS97 [20] program. The full matrix least square refinement using SHELXL97 program was used to include the position of all non hydrogen atoms. The thermal parameters for each atom were assigned a value of 0.05 (U’s) in the initial stage and refinement was followed. The initial scale factor was pegged at 1.0. Thereafter the anisotropic refinement for a few cycles of full matrix least square was continued. At this stage the positions of all hydrogen’s were geometrically fixed at calculated positions and they were allowed to ride on the corresponding non hydrogen atoms. The minimum and maximum value of residual electron density was -0.39, 0.38 e. Å-3 and the final R-factor were 0.061. Crystallographic data of the compound is summarized in Table 1.
| Parameter | Value |
|---|---|
| Formula | C14H14Cl2N2O3 |
| Formula weight | 329.17 |
| Crystal system | Triclinic |
| Space group | P -1 |
| T (K) | 295 (2) |
| a (Å) | 7.550 (3) |
| b (Å) | 9.426 (3) |
| c (Å) | 11.547 (3) |
| α (º) | 103.198 (4) |
| β (º) | 98.095 (4) |
| γ (º) | 104.099 (4) |
| V (Å3) | 759.0 (5) |
| Z | 2 |
| Dx (g cm-3) | 1.440 |
| F(000) | 340 |
| μ (mm-1) | 0.44 |
| Crystal size (mm) | 0.35 0.30 0.25 |
| Θ range (º) | 2.3–19.20 |
| hkl range | -7 ≤ h ≤ 6 -8 ≤ k ≤ 8 -10 ≤ l ≤ 10 |
| Reflections Collected | 4647 |
| Unique (Rint) | 1230 (0.063) |
| With [I>2σ(I)] | 905 |
| Number of parameters | 191 |
| R(F) [I>2σ(I)] | 0.063 |
| wR(F2) [I>2σ(I)] | 0.189 |
| R(F) [all data] | 0.061 |
| wR(F2) [all data] | 0.189 |
| Goodness of fit | 1.05 |
| Max/min ÃÆâÃâÃâ Ãâââ¬Â ρ (e Å-3) CCDC NO |
0.38/-0.39 |
Table 1 Crystal data, data collection and structure refinement.
Results and Discussion
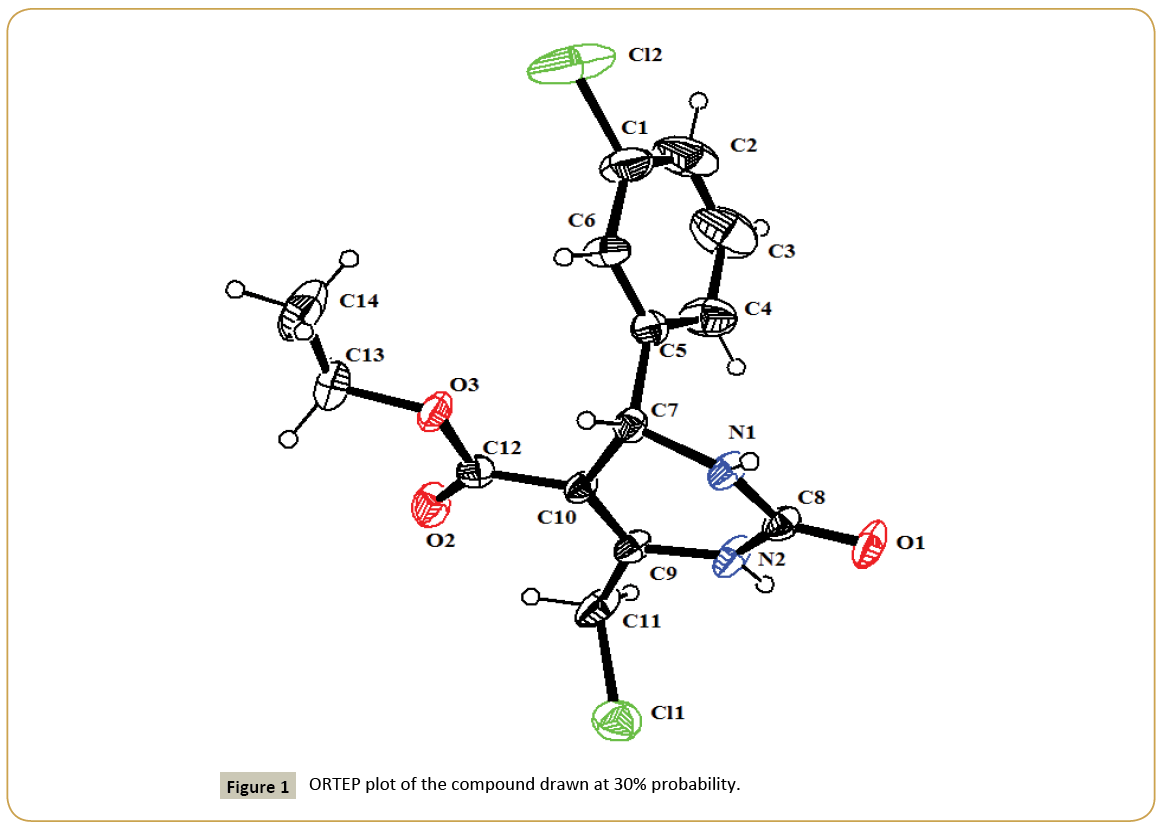
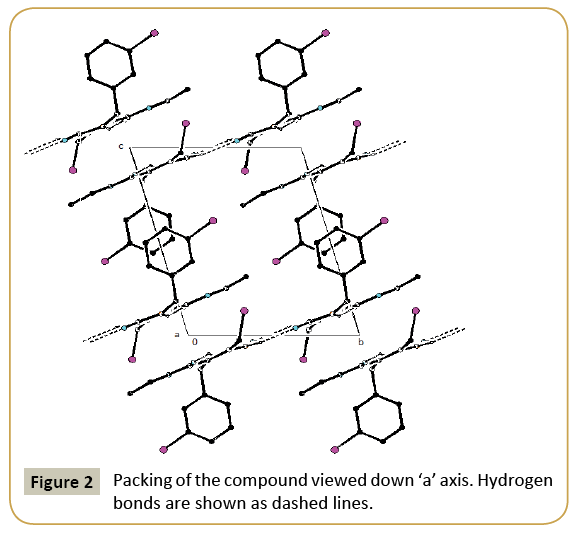
Figure 1 shows the ORTEP plot of the molecule drawn at 30% probability ellipsoid level with atom numbering scheme. Figure 2 shows the packing of compound viewed down ‘a’ axis. The geometric parameters of the title molecule (Figure 1) agree well with reported similar structure [21-23]. The chlorophenyl ring makes a dihedral angles of 86.25(4)° with the tetrahydropyrimidine ring. Table 2 summarizes the selected geometrical parameters of the compound. The molecular structure is stabilized by weak intra-molecular C-H···O interaction. The atom O2 is acting as potent acceptor for C11-H11B…O2 hydrogen bond in which atom C11 donates a proton. The interaction C11-H11B...O2 generates a sixmembered ring with S(6) graph set motif [24]. The crystal packing of the molecule is controlled by weak N-H…O interactions. All non-hydrogen atoms were refined anisotrophically and hydrogen atoms are fixed using riding modal. Two NH with oxygen atoms and one CH with oxygen atom are involved in non bonded interactions and possible hydrogen bonds are given in Table 3. In the crystal structure of molecule is interlinked via C11-H11B… Crystallographic data (excluding structure factors) for the structural analysis has been deposited with the Cambridge Crystallographic Data Centre, No. CCDC-848738. Copies of this information may be obtained free of charge from: The Director, CCDC, 12 Union Road, Cambridge, CB2 1EZ, UK. Fax: +44(1223)336-033, E-mail: deposit@ccdc.cam.ac.uk or web: www.ccdc.cam.ac.uk.
| Bond | Bond Length | Bond | Bond Length |
|---|---|---|---|
| C1—C2 | 1.354 (4) | C12—O2 | 1.205 (7) |
| C9—C10 | 1.331 (8) | C12—O3 | 1.329 (7) |
| C1—C6 | 1.381 (3) | C5—C6 | 1.358 (9) |
| C9—N2 | 1.394 (7) | C13—C14 | 1.452 (0) |
| C1—Cl2 | 1.723 (9) | C5—C7 | 1.526 (8) |
| C9—C11 | 1.483 (8) | C13—O3 | 1.460 (7) |
| C2—C3 | 1.351 (3) | C7—N1 | 1.482 (7) |
| C10—C12 | 1.487 (9) | C7—C10 | 1.503 (8) |
| C11—Cl1 | 1.760 (7) | C8—O1 | 1.228 (7) |
| C3—C4 | 1.377 (2) | C8—N1 | 1.333 (7) |
| C4—C5 | 1.364 (0) | C8—N2 | 1.357(8) |
| C2—C1—C6 | 122.3 (9) | C2—C1—Cl2 | 117.9 (1) |
| C9—C10—C7 | 120.6 (5) | C12—C10—C7 | 117.7 (6) |
| C6—C1—Cl2 | 119.8 (1) | ||
| C9—C11—Cl1 | 112.0 (5) | ||
| C3—C2—C1 | 117.8 (9) | ||
| C2—C3—C4 | 121.6 (1) | ||
| O2—C12—O3 | 122.4 (6) | ||
| C5—C4—C3 | 119.6 (8) | ||
| O2—C12—C10 | 126.9 (6) | ||
| O3—C12—C10 | 110.7 (6) | ||
| C14—C13—O3 | 107.9 (6) | ||
| C6—C5—C4 | 119.9 (7) | ||
| C6—C5—C7 | 120.0 (7) | ||
| C4—C5—C7 | 120.1 (6) | ||
| C5—C6—C1 | 118.8 (9) | ||
| N1—C7—C10 | 108.6 (5) | ||
| N1—C7—C5 | 110.7 (5) | ||
| C10—C7—C5 | 113.7 (5) | ||
| C8—N1—C7 | 124.1 (5) | ||
| O1—C8—N1 | 123.0 (6) | ||
| O1—C8—N2 | 121.3 (6) | ||
| N1—C8—N2 | 115.6 (6) | ||
| C8—N2—C9 | 123.8 (5) | ||
| C10—C9—N2 | 119.3 (5) | ||
| C10—C9—C11 | 127.9 (6) | ||
| N2—C9—C11 | 112.7 (5) | ||
| C12—O3—C13 | 116.9 (5) | ||
| C9—C10—C12 | 121.4(6) |
Table 2 Selected geometrical parameters (Å, ≡) with su’s in parentheses
| D-H...A | D-H | H...A | D...A | DHA |
|---|---|---|---|---|
| C11-H11B…O2 | 0.97 | 2.10 | 2.853(1) | 134 |
| N1-H1...O2i | 0.86 | 2.31 | 3.117(3) | 157 |
| N2-H2...O1ii | 0.86 | 2.06 | 2.907(1) | 170 |
Symmetry Equivalent position: (i) 1+x, y, z (ii) -x, -1-y, -z.
Table 3 Non-Bonded interactions and possible hydrogen bonds ( Å, ≡).
O2, N1-H1...O2 i and N2-H2...O1 ii hydrogen bonds to form R12 (6) ring motifs [17] which play a role in stabilizing the crystal structure. These sets of ring motifs are then linked into intermolecular hydrogen bonds.
Conclusion
Derivatives of Dihydropyrimidines exhibit a variety of medicinal properties by serving as antiviral, antitubercular, antimicrobial agents. The molecular structure of the compound is stabilized by weak intra-molecular C-H...O type of hydrogen bond. The crystal packing is controlled by weak inter-molecular N-H...O, interactions. Presence of inter and intra molecular hydrogen bonds in the title compound shows that, the derivative exhbit wide range of biological activities.
References
- Atwal KS, Rovnyak GC, Schwartz J, Moreland S, Hedberg A, et al. (1990) Dihydropyrimidine calcium channel blockers: 2-heterosubstituted 4-aryl-1,4-dihydro-6-methyl-5-pyrimidinecarboxylic acid esters as potent mimics of dihydropyridines. J Med Chem 33: 1510-1515.
- Kappe CO (1998) 4-Aryldihydropyrimidines via the Biginelli Condensation: Aza-Analogs of Nifedipine-Type Calcium Channel Modulators. Molecules 3: 1-9.
- Triggle DJ, Janis RA (1987) Calcium channel ligands. Annu Rev Pharmacol Toxicol 27: 347-369.
- Desai B, Sureja D, Naliapara Y, Shah A, Saxena AK (2001) Synthesis and QSAR studies of 4-substituted phenyl-2, 6-dimethyl-3, 5-bis-n-(substituted phenyl) carbamoyl-1,4-dihydropyridines as potential antitubercular agents. Bio Med Chem 9: 1993-1998.
- Kamaljit S, Kawaljit S, Baojie W, Scott F, Kelly C (2011) Facile transformation of Biginelli pyrimidin-2(1H)-ones to pyrimidines. In vitro evaluation as inhibitors of Mycobacterium tuberculosis and modulators of cytostatic activity. Bio Med Chem Let 46: 2290-2294.
- Atul DB, Kartik BV, Ketan BP, Kiran SN (2012) Synthesis of 1,2,3,4-tetrahydro pyrimidine derivatives as an antimicrobial agent. J Chem Pharm Res 4: 2972-2978.
- Bhuiyan MD, Rahman KM, Hossain MD, Rahim A, Hossain MI, et al. (2006) Synthesis and antimicrobial evaluation of some new thienopyrimidine derivatives. Acta Pharm 56: 441-450.
- Nitinkumar SS, Ravi SL, Imtiyaz AMK (2009) Synthesis and antimicrobial activity of some novel thienopyrimidine and triazolothienopyrimidines. J Chem Sci 121: 301-307.
- Sharma P, Rane N, Gurram VK (2004) Synthesis and QSAR studies of pyrimido[4,5-d]pyrimidine-2,5-dione derivatives as potential antimicrobial agents. Bioorg Med Chem Lett 14: 4185-4190.
- Wael AE, Omar MA, Rihana AZ, Asem AM and Abdul Rehman AH (2012) Synthesis and antimicrobial activity of new Substituted thienopyrimidines, their tetrazolyl and sugar derivatives. Act Pol Pharm 69: 439-447.
- Gillespie RJ, Cliffe IA, Dawson CE, Dourish CT, Gaur S, et al. (2008) Antagonists of the human adenosine A2A receptor. Part 3: Design and synthesis of pyrazolo[3,4-d]pyrimidines, pyrrolo[2,3-d]pyrimidines and 6-arylpurines. Bioorg Med Chem Lett 18: 2924-2929.
- Aurelio O, Ramon M, Beatriz L, Roberto O (2008) Novel 2-(4-methylsulfonylphenyl) pyrimidine derivatives as highly potent and specific COX-2 inhibitors. Bio Med Chem 16: 2183-99.
- Falcão EP, de Melo SJ, Srivastava RM, Catanho MT, Do Nascimento SC (2006) Synthesis and antiinflammatory activity of 4-amino-2-aryl-5-cyano-6-{3- and 4-(N-phthalimidophenyl)} pyrimidines. Eur J Med Chem 41: 276-282.
- Aleem G, Ying Z, Sudhir R, Michael AI, Bryan CD (2010) Design, synthesis and evaluation of 2-amino-4-m-bromoanilino-6-arylmethyl-7 H-pyrrolo [2,3 d]pyrimidine’s as tyrosine kinase inhibitors and antiangiogenic Agents. Bio Med Chem 18: 5261-5273.
- Parveen H, Hayat F, Salahuddin A, Azam A (2010) Synthesis, characterization and biological evaluation of novel 6-ferrocenyl-4-aryl-2-substituted pyrimidine derivatives. Eur J Med Chem 45: 3497-3503.
- Fuchun X, Hongbing Z, Lizhi Z, Liguang L, Youhong H (2009) Synthesis and biological evaluation of novel 2,4,5-substituted Pyrimidine derivatives for anticancer activity. Bio Med Chem Let 19: 275-278.
- Tangeda SJ, Garlapati A (2010) Synthesis of new pyrrolo[2,3-d]pyrimidine derivatives and evaluation of their activities against human colon cancer cell lines. Eur J Med Chem 45: 1453-1458.
- Akqun H, Karamelekogiu I, Berk B, Kurnaz I, Saribiyik G, et al. (2012) Synthesis and antimycobacterial activity of some phthalimide derivatives. Bioorg Med Chem 20: 4149-4154.
- Sheldrick GM (1996) SADABS, University of Gottingen, Germany.
- Sheldrick GM (1997) SHELXS97 & SHELXL97, University of Gottingen, Germany.
- Yuvaraj H, Sundaramoorthy S, Velmurugan D, Kalkhambkar RG (2010) Ethyl 4-(3-bromo-phen-yl)-6-methyl-2-oxo-1,2,3,4-tetra-hydro-pyrimidine-5-carboxyl-ate. Acta Crystallogr Sect E Struct Rep Online 66: o3325.
- Nayak SK, Venugopala KN, Chopra D, Govender T, Kruger HG (2009) Ethyl 4-(4-hydroxyphenyl)-6-methyl-2-oxo-1,2,3,4-tetrahydropyrimidine-5-carboxylate monohydrate. Acta Cryst E65: o2502.
- Bharanidharan S, Saleem H, Gunasekaran B, Padusha MS, Suresh M (2014) Crystal structure of ethyl 6-(chloro-meth-yl)-4-(4-chloro-phen-yl)-2-oxo-1,2,3,4-tetra-hydro-pyrimidine-5-carboxyl-ate. Acta Crystallogr Sect E Struct 70: o1185-1186.
- Bernstein J, Davis RE, Shimoni L and Chang NL (1995) Patterns in Hydrogen Bonding: Functionality and Graph Set Analysis in Crystals. Angew Chem Int Ed Engl 34: 1555–1573.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences