Anion Guided Self-Assembling of Guaninium Cations
Babulal Das and Jubaraj B Baruah
Babulal Das and Jubaraj BB*
Department of Chemistry, Indian Institute of Technology Guwahati, Guwahati-781 039, Assam, India
- *Corresponding Author:
- Jubaraj BB
Department of Chemistry, Indian Institute of Technology Guwahati, Guwahati-781 039, Assam, India
Tel: +913612582311
Fax: +913612690762
E-mail: juba@iitg.ernet.in
Received Date: February 22, 2016; Accepted Date: February 29, 2016; Published Date: March 05, 2016
Abstract
Anions guide self-assemblies of ribbons or end-capped dimers of guaninium cations. Self-assemblies of several salts of guanine are compared with related guanine salts from literature. End-capped dimers of guaninium cations selfassembled by perchlorate or by dihydrogen-phosphate anions whereas sulphatebisulphate anions stabilizes infinite ribbon-like assemblies of cations. Formation of discrete, dimer, or ribbons of cations in guaninium salts depends on anions and water has less dominant role other than to modify environments of anions. This result is a contrast effect caused by aquation on self-assembilies of neutral guanine molecules.
https://matadorbet-giris.com https://grandpashabeti.com https://betiste.com https://bahsegelgirisi.com https://klasbahisgirisi.com https://jasminbeti.com https://hepsibahise.com https://dinamobetgirisi.com https://betvolegirisi.com https://betpark-girisi.com https://betlikegiris.com https://betboogirisi.com https://sultanbetegiris.com https://tulipbetgir.com https://padisahbetegir.com https://savoybettinge.com https://goldenbahisegiris.com https://maksibetgirise.com https://fenomengiris.com https://jojobetgiris.xyz https://tarafgiris.com
Keywords
Guaninium cations; Self-assembly; Anion-assisted assembly; End capped structures; Ribbon-like assembly
Background
Aquated or free anions are found to guide self-assemblies of guaninium cations. End capped dimeric sub-assemblies are held by perchlorate or dihydrogen-phosphate to form self-assemblies; whereas sulphate-bisulphate guides guaninium cations to form ribbon-like assemblies of cations. Though hydration causes large variation of self-assembling of guanine, self-assembling of guaninium cations is invariant of aquation of anions.
Introduction
Self-complementary multiple hydrogen bond sites present in guanine and derivatives, help them to self-assemble into various supramolecular assemblies such as dimers, ribbons, tapes or macrocycles [1,2]. These assemblies have shown potential to develop sensors, nano-materials [3-5]. Famous G-quartet (G = guanine) assemblies find applications as hydrogels [6-9]. These GG pairs mainly assemble as ribbon or as part of cyclic G quartet in solid-state have applications in molecular electronics [10,11]. In addition, guanine derivatives form metal complexes [12-14]. Guanine nucleobases self-assemble to form four different type guanine-guanine pairs in DNA or RNA [15-18] which are shown in Scheme 1. Compartively, guanine has more numbers of ways to form self-assemblies than N-substituted guanines. However, poor solubility of guanine in common solvent stands as an obstacle to carry out structural study unsubstituted guanine, hence comparatively lesser structural study as compared to other base counterparts such as adenine have been carried out [19-21]. Broomhead [22] established structure of guanine hydrochloride monohydrate by X-ray in 1951, which followed structural study on salts such as nitrate [23] and phosphate and dihydrogenphosphate [24]. These (Scheme 1) reports are available in literature as segmented structural report and have not been made systematic; hence we set to study supramolecular architectures of structures of salts with various oxyanions. Lattice water molecules seldom participate in base pairing [25] but presence of single water molecule considerably influences guanine-guanine pairing patterns [26]. Keeping in mind the importance of such systems in crystal engineering [27] our focus in this study is on (i) protonation of guanine by varying tetrahedral anion from monobasic to tribasic acids, (ii) different self-assemblies of guanine or anion assisted assemblies in their protonated form, and (iii) role of lattice hydrates in assemblies.
Experimental Section
Synthesis and characterisation
Solution of guanine (G, 1 mmol) in water (5 ml) was reacted independently with 1 ml of acid, perchloric (70%) or sulphuric (98%) or phosphoric (85%) acid, respectively which afforded their salt 1, 2 or 3. Colorless solution obtained in each case was allowed to evaporate slowly at ambient temperature. After one week, single crystals suitable for X-ray crystallography were harvested. Salt [(HG)(ClO4)]1.5H2O 1: Yield, 48%. Elemental analysis calculated for C5H9ClN5O7.5, C, 20.37; H, 3.05; N, 23.76%; found C, 20.31; H, 3.04; N, 23.84%. IR (KBr, cm-1): 3436 (w), 3377 (m), 3167 (m), 3102 (m), 2922 (m), 1711 (s), 1654 (s), 1607 (w), 1388 (s), 1144 (w), 1111 (m), 1084 (s). Molar conductance: 307.0 Scm2mol-1 in water. Salt [(HG)2(HSO4)2(SO4)]2H2O (2): Yield, 45%. Elemental analysis calculated for C20H30N20O18S3, C, 25.67; H, 3.21; N, 29.95%; found C, 25.60; H, 3.14; N, 30.02%. IR (KBr, cm- 1): 3336 (bs), 3104 (bs), 2907 (w), 1697 (s), 1654 (m), 1615 (m), 1375 (s), 1073 (s). Molar conductance: 524.0 Scm2mol-1 in water. Thermal analysis: decomposition range ~ 80-175 °C (loss of two water molecules of crystallization). Salt [(HG)(H2PO4)]H2O (3): Yield, 52%. Elemental analysis calculated for C5H10N5O6P, C, 22.46; H, 3.74; N, 26.20%; found C, 22.33; H, 3.69; N, 25.95%. Selected IR data (KBr, cm-1): 3473 (w), 3333 (w), 3178 (w), 3139 (s), 2922 (w), 1672 (s), 1615 (s), 1382 (m), 1099 (m), 1047 (s), 965 (m). Molar conductance: 168.0 Scm2mol-1 in water. Thermal analysis: decomposition range: ~70-135 °C (loss of water molecules).
Physical measurements
Infrared spectra (KBr pellets) of salts 1-3 were recorded with a Thermo iS10 FTIR spectrophotometer in 4000-400 cm-1 spectral region. Elemental analyses were performed with a Perkin Elmer 2400 series micro analytical analyzer. Elico conductivity meter, model CM 180, was used to determine molar conductance.
X-ray structural studies
Single crystal diffraction data for 1-3 were collected at 296 K with Mo Kα radiation (λ = 0.71073 Å) using a Bruker Nonius SMART APEX diffractometer equipped with graphite monochromator and Apex CCD camera. SMART software (v 2.1-4) was used for indexing and unit cell parameters. Data reduction and cell refinement were performed using SAINT software and the space groups of these crystals were determined from systematic absences by XPREP. The structures were solved by direct methods and refined by full-matrix least-square calculations using SHELXTL software. All the non-hydrogen atoms were refined in anisotropic approximation against F2 of all reflections. Hydrogen atoms attached to nitrogen atoms of guanine were located in difference Fourier synthesis maps, and refined with isotropic displacement coefficients. Crystal parameters are summarized in Table 1.
| Compound No. | Salt 1 | Salt 2 | Salt 3 (reported earlier31) |
|---|---|---|---|
| Formula | C10H18Cl2N10O15 | C20H30N20O18S3 | C5H10N5O6P |
| Mr | 589.24 | 934.82 | 267.15 |
| Crystal system | Monoclinic | Triclinic | Monoclinic |
| Space group | Cc | P-1 | P21 /n |
| a (Å) | 4.9051(3) | 6.4530(5) | 4.5542(2) |
| b (Å) | 46.567(3) | 13.3515(9) | 12.5955(5) |
| c (Å) | 9.7185(6) | 20.9920(14) | 18.2991(7) |
| α (deg) | 90.00 | 77.048(4) | 90.00 |
| β (deg) | 96.646(4) | 81.672(4) | 92.606(2) |
| γ (deg) | 90.00 | 86.203(4) | 90.00 |
| V (Å3) | 2204.9(2) | 1742.9(2) | 1048.60(7) |
| Z | 4 | 2 | 4 |
| Dcalc (gcm-3) | 1.775 | 1.781 | 1.692 |
| μ (mm-1) | 0.393 | 0.324 | 0.292 |
| F(000) | 1208 | 964 | 552 |
| Total no. of reflns | 15906 | 19714 | 10860 |
| Independent reflns | 3676 | 5853 | 1833 |
| θrange | 0.87 – 24.98 | 1.00 – 25.00 | 1.96 - 25.00 |
| Ranges (h, k, l) |
-5 ≤ h ≤ 5 -54 ≤ k ≤ 54 -11 ≤ l ≤ 11 |
-7 ≤ h ≤ 7 -15 ≤ k ≤ 14 -24 ≤ l ≤ 22 |
-5 ≤ h ≤ 5 -14 ≤ k ≤ 14 -21 ≤ l ≤ 21 |
| Completeness to 2θ (%) |
99.2 | 95.1 | 98.9 |
| Data /restraints/ parameters | 3676 / 6 / 358 | 5853 / 0 / 562 | 1833 / 0 / 182 |
| GOF (F2) | 0.820 | 1.195 | 1.070 |
| R1, wR2 [I > 2σ(I)] |
0.0612, 0.1648 | 0.0769, 0.1925 | 0.0328, 0.0899 |
| R1, wR2 (all data) |
0.0862, 0.1956 | 0.1112, 0.2138 | 0.0389, 0.0944 |
| Largest diff peak / hole (e Å-3) | 0.392 / -0.650 | 0.907 / -0.587 | 0.272 / -0.422 |
Supporting information: Crystallographic Information files of the salts 1-3 are deposited to Cambridge Crystallographic database and they have CCDC Nos. 1014792-1014794.
Table 1: Crystal parameters for the structures 1-3.
Results and Discussion
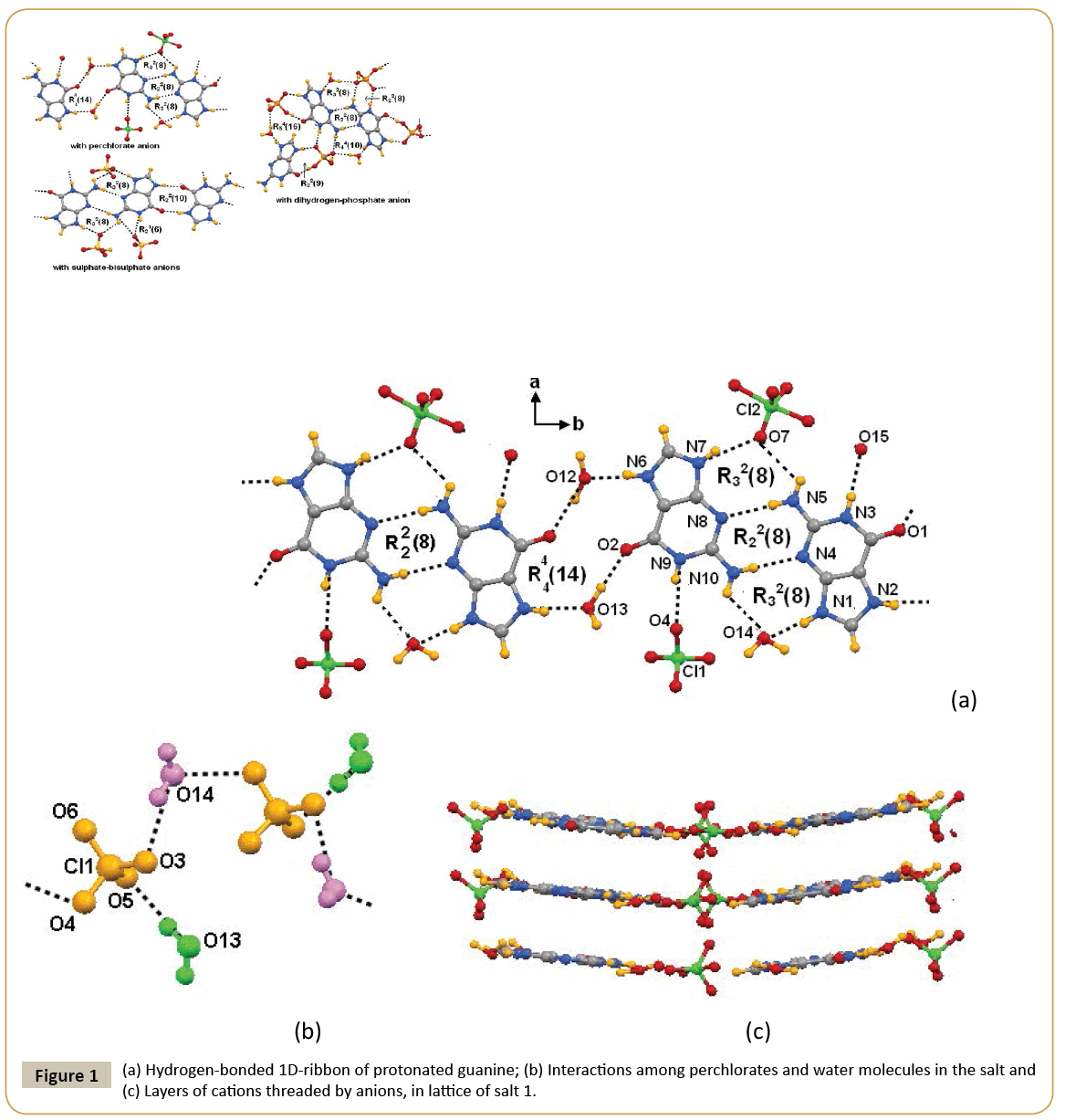
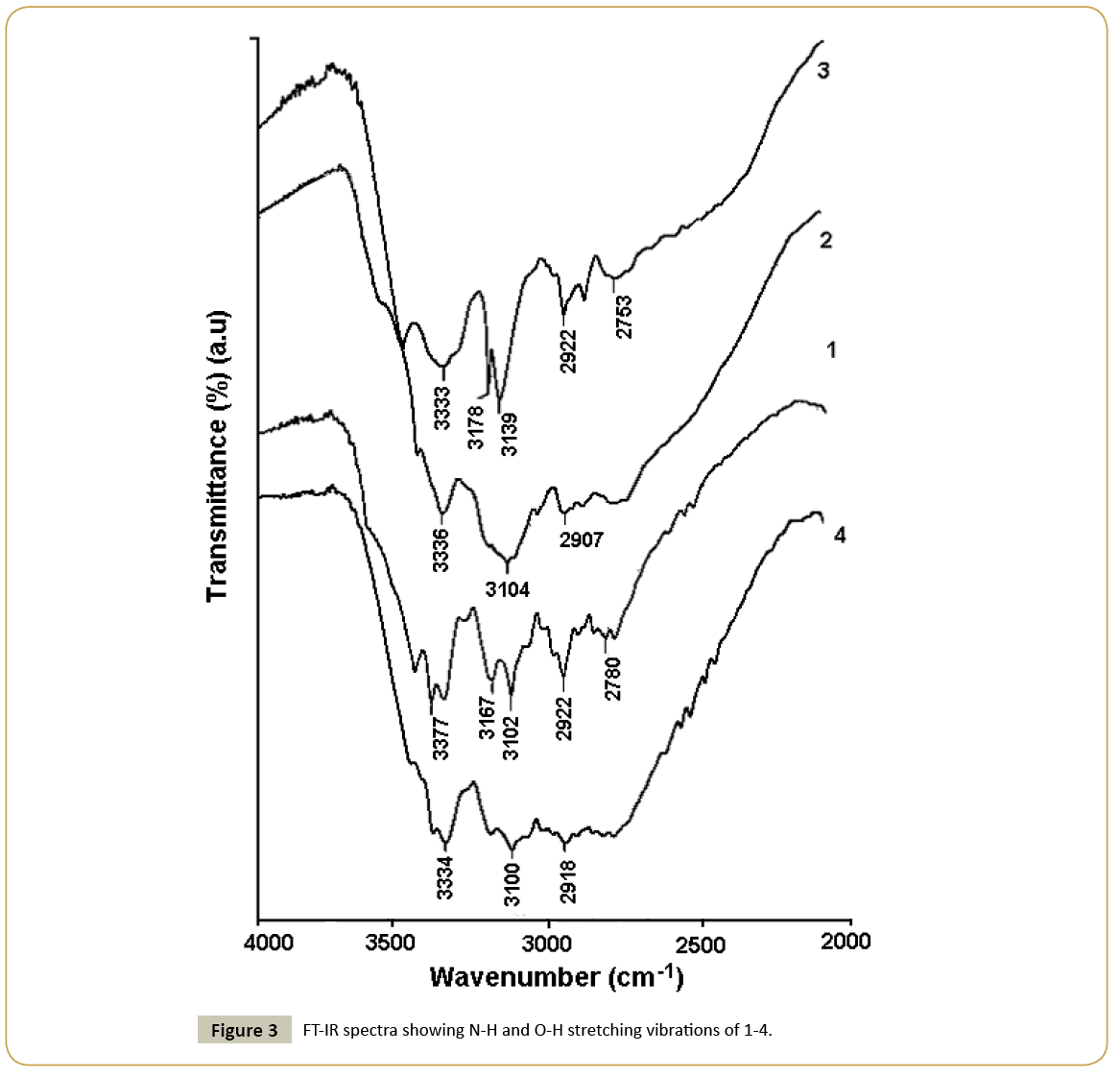
Guanine was treated with different oxy-acids possessing tetrahedral anion in aqueous medium with an anticipation to generate hydrated assemblies of salts. Treatment of guanine (G) with perchloric, sulphuric or phosphoric acid in water afforded respective salt (HG)(ClO4)·1.5H2O (1), (HG)4(HSO4)2(SO4)·2H2O (2) or (HG)(H2PO4)·H2O (3). It may be noticed that the hydration in each salt is different. The dihydrogen-phosphate salt was reported earlier [24] but we have redone the structure so as to make a comparison on crystals obtained from three different acids under similar conditions. Crystal structure of guaninium-perchlorate (1) consists of two units of guaninium cation [HG]+ and perchlorate anion, and four lattice water molecules. In the perchlorate salt 1, protonation by perchloric acid took place at conventional site as observed earlier [28]. In the crystal structure, one oxygen atom of perchlorate is found to be disordered. The electron density of disordered atom is distributed over two crystallographically equivalent positions. Due to flexible interacting ability among supramolecular components possibly allowed the crystallographic disorder in perchlorate anion [29]. Guaninium cations form dimer through Watson-Crick edge by interacting through N-H···N interactions between two symmetry independent cations form R22(8) motif [30,31] (Figure 1a). Due to interaction of such subassemblies of cations two lattice water molecules results in an extended one-dimensional ribbon along crystallographic c-axis cations which posseses R44(14) motif as illustrated in Figure 1a. The perchlorate anions and lattice water molecules form onedimensional chain as shown in Figure 1b. In lattice, adjacent guanine-perchlorate ribbons reside in a parallel manner, holding perchlorate anions from either side (Figure 1c). Distances between such planes are ~ 3.2 Å. These chains provide connecting points to keep the dimeric pairs of cations at two ends to form grid like arrangements of cations. On the other hand depending on host, aquated perchlorate anions were shown form hexameric cyclic hydrogen bonded assembly having appearance of open-book [32,33]. Hence, present example shows stabilisation of aquated assemblies of perchlorate anions by dimeric guaninium cations. The structure amy be also describe alternatively as assemly of end capped dimeric cations capped by perchlorate ions through a R32(8) motif on one side and water molecules froming on other sides and such units held by R44(14) motifs (Table 2). Single crystals obtained from reaction of guanine with sulphuric acid in water, had a composition (HG)4(HSO4)2(SO4)·2H2O (2). This result is different from earlier report as heating aqueous solution of guanine with sulpuric acid gave the guaninium salt (HG)2SO4.2.5H2O [31]. Hence, it clearly points out that variation of recation conditions significantly affects the deprotonation of the acid. The salt 2 is comprised of both sulphate and bisulphate anion. The asymmetric unit has four [HG]+ cations, two bisulphate anions, one sulphate anion and two lattice water molecules. Anhydrous form of guanine salt having sulphate and bisulphate was prepared earlier by dehydration of corresponding hydrated salt [31]. Anhydrous salt had a similar ribbon-like assembly of cations as observed in the hydrated salt. In hydrated anions holds the ribbons, whereas in anhydrous form they are held directly by the anions. Thus, role of water in presence of sulphate-bisulphate anion has little to do to change the assembling of cations. Among the four [HG]+ cations present in the asymmetric unit, two are symmetry independent, the symmetry equivalent pair form one-dimensional planar ribbon through R22(10) motifs along crystallographic a-axis as shown in Figure 2a. Hydrogen bond parameters are listed in Table 2. Close look at crystal structure of 2 shows that cationic guaninium ribbons are aligned parallel separated by a distance of 3.24 Å (Figure 2b). Sulphate and bisulphate anions lie on two sides of ribbons. Sulphate anions are held by bifurcated hydrogen bonds with two N-H bonds, forming a R21(6) motifs on one side and R32(8) on another side. On the other hand bisulphate are held to cations by R32(8) motif on one side and by N-H···O interactions (Figure 2a) on another side. Anions and lattice water molecules form infinite anion-water cluster as shown in Figure 2c. Common feature between the guaninium sulphate salts reported by others and us is guaninium cations showed same type of cationic ribbon based on cyclic hydrogen bonded R22(8) and R22(10) motifs. Phosphoric acid reacted with guanine to form a salt with a composition (HG)(H2PO4)·H2O (3), structure of which was reported earlier [24]. It posses end capped cationic dimeric assemblies bound to dihydrogen phosphate anions and water molecules through R22(8) and R44(10) hydrogen bonded motifs (Figure 2c). These end capped structures are linked to each other through R22(9) and R54(16) type hydrogen bond motifs. On the other hand, nitrate salt of guanine (H2G) (NO3)2·2H2O (4) reported [23] earlier was also confirmed by us that same is also formed under ordinary condition on reaction of guanine with nitric acid. It is comprised of diprotonated guanine forming planar sheet like assembly with nitrate anions and lattice water molecules help in stacking. Infra-red spectra of salts 1-4 show stretching vibrations appearing in the range of 3436-3472 cm-1 (Figure 3) from hydrogen bonded water molecules [34,35]. The N-H stretching vibrations appear in range 3100-3377 cm-1 but stretching frequencies differ with anion; suggests different hydrogen bonding environment for the cationic part in each case. In these regions, guanine salts containing perchlorate and dihydrogen-phosphate anions exhibit sharp stretching frequencies, while marked broadening peaks appeared for the nitrate and sulphate-bisulphate containing guanine salts. This is attributed to difference in hydrogen bonds associated with dimer or infinite ribbon of protonated guanine molecules surrounded by anions. Each salt also exhibits characteristic sharp stretching vibrations for corresponding anion. Generally IR-stretching of perchlorate ions is guided by environment around it, perchlorate anion in symmetric environment or free anion shows one single stretching around 1150 cm-1, however we observed three symmetric shows signals at 1143 cm-1, 1111 cm-1 and 1083 cm-1. This supports that the perchlorate is under influence of strong hydrogen bond as illustrated in Figure 2a. In case of sulphate absorption due to S=O appears at 1073 cm-1. Whereas, biphosphate shows strong absorptions due to P=O stretching at 1099 cm-1, 1047 cm-1 and 965 cm-1 respectively whereas the nitrate salts shows N=O stretching at 1384 cm-1.
| Bond (symmetry) | dD-H (Å) | dHÃÆâÃâÃâ Ãâââ¢ÃÆâÃâÃâ Ãâââ¢ÃÆâÃâÃâ Ãâââ¢A (Å) | dDÃÆâÃâÃâ Ãâââ¢ÃÆâÃâÃâ Ãâââ¢ÃÆâÃâÃâ Ãâââ¢A (Å) | <D-HÃÆâÃâÃâ Ãâââ¢ÃÆâÃâÃâ Ãâââ¢ÃÆâÃâÃâ Ãâââ¢(Å) | |
|---|---|---|---|---|---|
| 1 | N(1)-H(1A)···O(14) | 0.86 | 1.89 | 2.745(8) | 171 |
| N(2)-H(2)···O(13)i | 0.86 | 1.85 | 2.700(8) | 167 | |
| N(3)-H(3)···O(15) | 0.86 | 1.97 | 2.834(9) | 177 | |
| N(5)-H(5A)···N(8)ii | 0.86 | 2.24 | 3.102(8) | 175 | |
| N(6)-H(6A)···O(12) | 0.86 | 1.80 | 2.656(9) | 170 | |
| N(7)-H(7)···O(7)iii | 0.86 | 1.99 | 2.827(12) | 163 | |
| N(9)-H(9)···O(4) | 0.86 | 2.08 | 2.912(7) | 161 | |
| N(10)-H(10A)···N(4)iii | 0.86 | 2.16 | 3.015(8) | 177 | |
| O(12)-H(12A)···O(1)iv | 0.97(12) | 1.84(15) | 2.741(8) | 154(18) | |
| O(13)-H(13B)···O(2) | 0.97(5) | 1.83(5) | 2.799(7) | 179(7) | |
| 2 | N(1)-H(1N)···O(16)v | 0.86 | 1.89 | 2.735(5) | 168 |
| N(2)-H(2N)···O(2)vi | 0.95(5) | 1.75(5) | 2.694(5) | 171(4) | |
| N(4)-H(4N)···O(11) | 0.86 | 1.99 | 2.827(5) | 164 | |
| N(5)-H(5NB)···N(9) | 0.86 | 2.21 | 3.060(6) | 173 | |
| N(6)-H(6N)···O(7) | 0.86 | 1.83 | 2.675(5) | 168 | |
| N(7)-H(7N)···O(1)vii | 0.86 | 1.90 | 2.757(5) | 172 | |
| N(8)-H(8N)···O(5)v | 0.86 | 2.04 | 2.840(5) | 155 | |
| N(10)-H(10A)···N(3) | 0.86 | 2.22 | 3.079(6) | 175 | |
| N(11)-H(11N)···O(8)ix | 0.86 | 1.82 | 2.659(5) | 166 | |
| N(12)-H(12N)···O(4)viii | 0.99(8) | 1.78(8) | 2.762(5) | 174(9) | |
| N(13)-H(13N)···O(17) | 0.86 | 2.03 | 2.873(5) | 168 | |
| N(15)-H(15A)···N(19)ix | 0.86 | 2.27 | 3.125(6) | 172 | |
| N(15)-H(15B)···O10 | 0.86 | 2.12 | 2.949(5) | 163 | |
| N(16)-H(16N)···O(9)ix | 0.86 | 1.78 | 2.610(6) | 163 | |
| N(17)-H(17N)···O(3)viii | 0.95(4) | 1.70(4) | 2.655(5) | 177(4) | |
| N(18)-H(18N)···O13 | 0.86 | 1.99 | 2.819(5) | 160 | |
| N(20)-H(20A)···N(14)ix | 0.86 | 2.21 | 3.066(6) | 174 |
Symmetry codes: (i) 1+x, y, -1+z; (ii) -1+x, y, z; (iii) 1+x, y, z; (iv) -1+x, y, 1+z; (v) 1-x, 1-y, -z; (vi) x, 1+y, z; (vii) x, -1+y, z; (viii) 1-x, -y, 1-z; (ix) 1-x, 1-y, 1-z.
Table 2: Selected hydrogen-bond parameters for the salts 1-2.
Conclusion
Hydrated salts of guanine salts of mono-, di-, tri-basic acids have shown highly anion dependent self-assemblies to provide scope for formation of ribbon or end capped or discrete self-assemblies of guaninium cations. On the other hand, planar nitrate ion acts as a template to stabilize discrete guaninium di-cations. Triple hydrogen bonded assemblies which generally contribute to insoluble nature of guanine, are absent in any of these structures of the salt studied. It is also observed that water molecules do not change the arrangements of cationic assemblies of guanine in hydrated or anhydrous form of salt, this result is advantageous over the structural changes caused by hydration in self-assemblies of neutral guanine molecules.
References
- Davis JT, Spada GP (2007) Supramolecular architectures generated by self-assembly of guanosine derivatives. Chem Soc Rev 36: 296-313.
- Spindler L, Fritzsche W (2012) Guanine quartets: Structure and application. RSC publishing, Cambridge, UK.
- Gottarelli G, Masiero S, Mezzina E, Pieraccini S, Rabe JP, et al. (2000) The self-assembly of lipophilic guanosine derivatives in solution and on solid surfaces. Chem Eur J 6: 3242-3248.
- Van LF, Verboom W, Shi XD, Davis JT, Reinhoudt DN (2004) Selective 226Ra2+ ionophores provided by self-assembly of guanosine and isoguanosine derivatives. J Am Chem Soc 126: 16575-16581.
- Neviani P, Sarazin D, Schmutz M, Blanck C, Giuseppone N, et al. (2010) Hierarchical formation of fibrillar and lamellar self-assemblies from guanosine-based motifs. J Nucleic Acids.
- Wong A, Ida R, Spindler L, Wu G (2005) Disodium guanosine 5'-monophosphate Self-associates into nanoscale cylinders at pH 8: A combined diffusion NMR spectroscopy and dynamic light scattering study. J Am Chem Soc 127: 6990-6998.
- Sreeni-vasachary N, Lehn JM (2005) Gelation-driven component selection in the generation of constitutional dynamic hydrogels based on guanine-quartet formation. Proc Natl Acad Sci USA 102: 5938-5943.
- Giorgi T, Grepioni F, Manet I, Mariani P, Masiero S, et al. (2002) Gel-like lyomesophases formed in organic solvents by self-assembled guanine ribbons. Chem Eur J 8: 2143-2152.
- Giorgi T, Lena S, Mariani P, Cremonini MA, Masiero S, et al. (2003) Supramolecular helices via self-assembly of 8-oxoguanosines. J Am Chem Soc 125: 14741-14749.
- Rinaldi R, Maruccio G, Biasco A, Arima V, Cingolani R, et al. (2002) Hybrid molecular electronic devices based on modified deoxyguanosines. Nanotechnology 13: 398-403.
- Murata T, Saito G, Nakamura K, Maesato M, Hiramatsu T, et al. (2013) Exploration of charge-transfer solids utilizing nucleobases: Nanoarchitectures by hydrogen-bonds in the ionic assemblies of guanine and TCNQ derivatives. Cryst Growth Des 13: 2778-2792.
- Gupta D, Nowak R, Lippert B (2010) Pt(II) complexes of unsubstituted guanine and 7-methylguanine. Dalton Trans 39: 73-84.
- Nagapradeep N, Sharma S, Verma S (2013) Ion channel-like crystallographic signatures in modified guanine-potassium/sodium interactions. Cryst Growth Des 13: 455-459.
- Mastropietro TF, Armentano D, Grisolia E, Zanchini C, Lloret F, et al. (2008) Guanine-containing copper(II) complexes: synthesis, X-ray structures and magnetic properties. Dalton Trans 514-520.
- Saenger W (1984) Principles of nucleic acid structures. Springer, Berlin.
- Kozma A, Ibanez S, Silaghi-Dumitrescu R, Miguel PJS, Gupta D, et al. (2012) 7-Methylguanine: protonation, formation of linkage isomers with trans-(NH3)2PtII, and base pairing properties. Dalton Trans 41: 6094-6103.
- Brown T, Hunter WN (1997) Non-Watson-Crick base associations in DNA and RNA revealed by single crystal X-ray diffraction methods: Mismatches, modified bases, and non-duplex DNA. Biopolymers 44: 91-103.
- Leontis NB, Westhof E (2001) Geometric nomenclature and classification of RNA base pairs. RNA 7: 499-512.
- Verma S, Mishra AK, Kumar J (2010) The many facets of adenine: coordination, crystal patterns, and catalysis. Acc Chem Res 43: 79-91.
- Das B, Baruah JB (2010) Protonated adenine and cytosine ribbons stabilized by dipicolinato metal frameworks. Cryst Growth Des 10: 3242-3249.
- Bendjeddou L, Cherouana A, Hadjadj N, Dahaoui S, Lecomte C (2009) Adeninium 3-carboxy-anilinium bis-(perchlorate) trihydrate. Acta Crystallogr 65E: 2303-2304.
- Broomhead JM (1951) The structures of pyrimidines and purines. IV. The crystal structure of guanine hydrochloride and its relation to that of adenine hydrochloride. Acta Crystallogr 4: 92-100.
- Bouchouit K, Benali-Cherif N, Benguedouar L, Bendheif L, Merazig H (2002) Guaninium dinitrate dehydrate. Acta Crystallogr 58E: o1397-o1399.
- Bendeif EE, Dahaoui S, Benali-Cherif N, Lecomte C (2007) Tautomerism and hydrogen bonding in guaninium phosphite and guaninium phosphate salts. Acta Crystallogr 63B: 448-458.
- Roitzsch M, Lippert B (2005) Structural precursor of the hemideprotonated guanine pair. Chem Commun 5991-5993.
- Abo-Riziq A, Crews B, Grace L, deVries MS (2005) Microhydration of guanine base pairs. J Am Chem Soc 127: 2374-2375.
- Aakeroy CB, Seddon KR (1993) The hydrogen bond and crystal engineering. Chem Soc Rev 22: 397-407.
- Hoxha K, Prior TJ (2013) Retention of crystallinity in bis(guaninium)sulfate hydrate upon partial and full dehydration. Solid State Sci 23: 102-108.
- Jang YH, Goddard III WA, Noyes KT, Sowers LC, Hwang S, et al. (2003) pKa Values of guanine in water: density functional theory calculations combined with Poisson-Boltzmann continuum-solvation model. J Phys Chem 107B: 344-357.
- Etter MC (1990) Encoding and decoding hydrogen-bond patterns of organic compounds. Acc Chem Res 23: 120-126.
- Bernstein J, Davis RE, Shimoni L, Chang NL (1995) Patterns in hydrogen bonding: functionality and graph set analysis in crystals. Angew Chem Int Ed 34: 1555-1573.
- Perez C, Muckle MT, Zaleski DP, Seifert NA, Temelso B, et al. (2012) Structures of cage, prism, and book isomers of water hexamer from broadband rotational spectroscopy. Science 336: 897-901.
- Phukan N, Baruah JB (2014) Polymorphs of 1-(5-Methylthiazol-2-yl)-3-phenylthiourea and various anion-assisted assemblies of two positional isomers. Cryst Growth Des 14: 2640-2653.
- Silverstein RM, Webster FX, Kiemle DJ (2005) Spectroscopic determination of organic compounds. 7th edn. Wiley: New York.
- Sheldrick GM (2008) A short history of SHELX. Acta Crystallogr 64A: 112-122.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences