Synthesis and Crystal Structure of 3-{4-[(4-(2-Oxo-2,3-dihydro-1H-benzimidazol- 1-yl)piperidin-1-yl)benzyl]}-2-phenylindole
J Guillon, Vincenzi M, Pinaud N, Ronga L, Rossi F, Savrimoutou S, Moreau S, Desplat V and Marchivie M
J Guillon1*, Vincenzi M1,2, Pinaud N3, Ronga L1, Rossi F2, Savrimoutou S1, Moreau S1, Desplat V4 and Marchivie M5
1Université de Bordeaux, UFR des Sciences Pharmaceutiques, INSERM U1212 / UMR CNRS 5320, Laboratoire ARNA, 146 rue Léo Saignat, F-33076 Bordeaux cedex, France
2Department of Pharmacy and CIRPeB, University of Naples “Federico II”, Via Mezzocannone, 16 80134 Naples, Italy
3Université de Bordeaux, ISM - CNRS UMR 5255, 351 cours de la Libération, F-33405 Talence cedex, France
4Université de Bordeaux, UFR des Sciences Pharmaceutiques, Laboratoire BMGIC INSERM U1035, 146 rue Léo Saignat, F-33076 Bordeaux cedex, France
5Université de Bordeaux, ICMCB CNRSUPR 9048, 87 Avenue du Docteur Schweitzer, 33608 Pessac cedex, France
- *Corresponding Author:
- Guillon J
Université de Bordeaux, UFR des Sciences Pharmaceutiques, INSERM U1212/UMR CNRS 5320, Laboratoire ARNA, 146 rue Léo Saignat, F-33076 Bordeaux cedex, France.
Tel: +33(0)557571652
E-mail: jean.guillon@u-bordeaux.fr
Received Date: March 14, 2016; Accepted Date: April 02, 2016; Published Date: April 07, 2016
Abstract
The X-ray crystal structure of 3-{4-[(4-(2-oxo-2,3-dihydro-1H-benzimidazol- 1-yl)piperidin-1-yl)benzyl]}-2-phenylindole, a compound that showed high cytotoxic potential against various leukemia cell lines, was established. The 3D spatial determination confirmed the structure in the solid state. It crystallizes in the triclinic space group P-1 with cell parameters a=9.572(7) Å, b=11.091(2) Å, c=15.976(4) Å, α=92.53(2), β=105.45(3), γ=94.65(3), V=1625.6(13) Å3 and Z=2. The crystal structure was refined to final values of R1=0.0638 and wR2=0.1800. The crystal packing is controlled by intermolecular hydrogen bonding and C-H ðÂÂœ‹ interactions. In addition, these new data could be further used to clarify the mechanism of action, implicating the antiproliferative effect of this new synthetic indole derivative on human leukemic cells.
Keywords
Single-crystal x-ray study; Indole derivative; Antiproliferative activity; R factor
Introduction
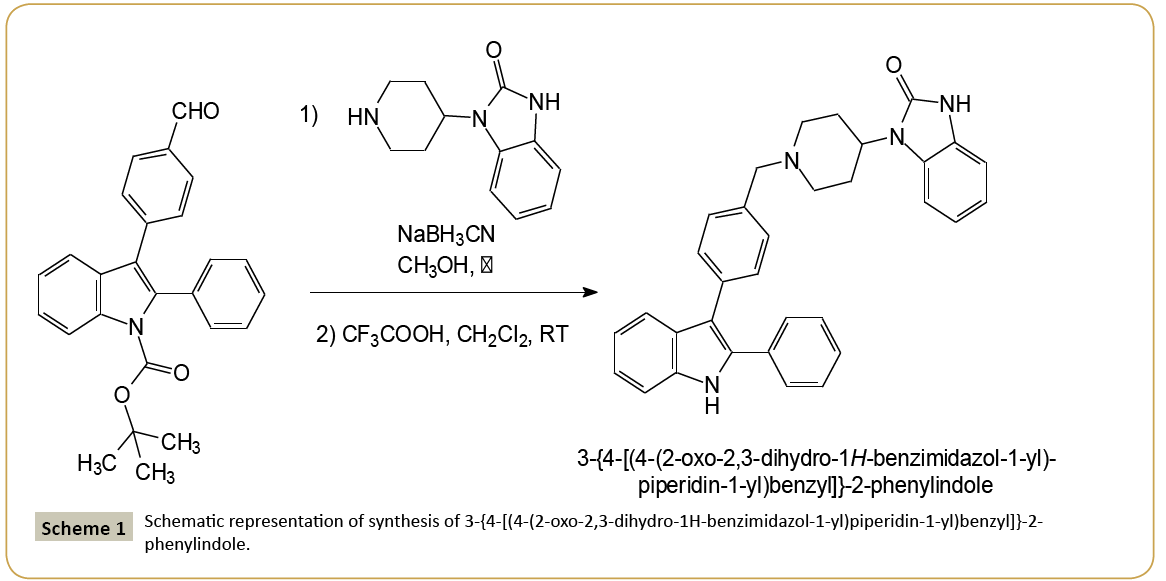
Acute leukemia is one of the most aggressive hematopoietic malignancies and is characterized by the abnormal proliferation of the immature cells and a premature block in lymphoid or myeloid differentiation. Adult acute leukemia has a poor prognosis due to a large number of relapses, because of treatment-related resistance mechanisms [1]. Therefore, there is an urgent need to find new therapeutics, which could led to the development of novel treatment strategies with less or minimal side effects. Heterocyclic compounds attracted a lot of attention because of its wide spread biological activities. Among them, the indole heterocyclic framework constitutes the basis of an important class of compounds possessing interesting biological activities [2-7]. These compounds have been reported to serve as key intermediates for the assembly of several heterocycles including anti-migraine agents, antiviral agents, antiparasitic agents, and antitumor agents [8-13]. In this last field, the discovery and development of novel therapeutic agents are one of the most important goals in medicinal chemistry. In the course of our work devoted to discover new compounds employed in the cancer chemotherapy, we previously identified four series of substituted heterocyclic pyrrolo[1,2-a]quinoxaline derivatives endowed with good activity towards the human leukemia cells [14-17]. In this context, and as an extension of our work on the development of new anticancer heterocyclic drugs, we decided to substitute our pyrrolo[1,2-a]quinoxaline pharmacophore by a bio-isostere heterocyclic analogue, i.e., the bioactive indole moiety. Thus, we report herein on the synthesis and structural characterization of a new analogue of these previously described compounds based on an indole scaffold, i.e., the 3-{4-[(4-(2-oxo- 2,3-dihydro-1H-benzimidazol-1-yl)piperidin-1-yl)benzyl]}-2- phenylindole. The synthesis of this new indole derivative is depicted in Scheme 1. Its preparation involved a reductive amination of tert-butyl 3-(4-formylphenyl)-2-phenyl-indole-1- carboxylate with 4-(2-ketobenzimidazolin-1-yl)piperidine using sodium cyanoborohydride (NaBH3CN), then followed by the cleavage of the Boc-protecting group of indole achieved using a trifluoroacetic acid solution in dichloromethane leading to the title compound. The cytotoxicity of this new compound was then evaluated against five different leukemia cell lines, including Jurkat and U266 (lymphoid cell lines), and K562, U937, HL60 (myeloid cell lines), as well as normal human peripheral blood mononuclear cells (PBMNCs) [14-17]. The preliminary biological results exhibited inhibitory activity in a submicromolar range against different leukemia cell lines, including Jurkat (lymphoid cell line), K562 and HL60 (myeloid cell lines) with IC50 of 8, 5 and 12 μM, respectively. This compound was also tested on activated (PBMNC + PHA) human peripheral blood mononuclear cells to evaluate its cytotoxicity on normal cells (IC50>50 μM). Indexes of selectivity (IS) were defined as the ratio of the IC50 value on the human mononuclear cells to the IC50 value on the different leukemia cell lines. Thus, the title compound shows an interesting index of selectivity >10 on the human myeloid leukemia cell lines K562. The present crystal structure determination will not only help us understand the detailed three-dimensional arrangement of the compound, which could be useful for designing new derivatives, but will also contribute to the structural database in which there are very few structures containing the indole skeleton. Moreover, solid-state data could be used to clarify the mechanism of action implicating this anti-proliferative derivative. on silica gel using ethyl acetate - cyclohexane (1/1) as eluents gave a white powder. To a cooled solution of this latter solid (0.5 mmol) in 5 mL of dichloromethane was added 10 mL of a 15% trifluoroacetic acid solution in dichloromethane. The mixture was stirred at room temperature for 24 h, then neutralized with 15 mL of a saturated aqueous solution of potassium carbonate and extracted with 3 × 10 mL of dichloromethane. The organic layer was washed with brine and dried with anhydrous sodium sulphate. The solvent was removed under reduced pressure. The crude residue was then purified by column chromatography using ethyl acetate - cyclohexane (1/1) as eluents gave a white powder of the title compound with melting point 169°C and yield 64%. Single crystals suitable for X-ray diffraction analysis were obtained by slow evaporation of the compound from a methanol-dichloromethane solution (3/7 – v/v) at +20°C. IR (KBr) 3403 and 3221 (NH), 1692 (CO). 1H NMR (CDCl3) δ: 9.36 (s, 1H, NH indole), 8.39 (s, 1H, NH benzimid.), 7.74 (d, 1H, J=7.80 Hz, H indole), 7.49-7.18 (m, 11H, H indole, H phenyl and H benzimid.), 7.13 (t, 1H, J=7.80 Hz, H indole), 7.12-7.05 (m, 4H, H phenyl and H benzimid.), 4.49-4.37 (m, 1H, CH pip.), 3.65 (s, 2H, CH2N), 3.17- 3.13 (m, 2H, CH2 pip.), 2.59-2.47 (m, 2H, CH2 pip.), 2.29-2.21 (m, 2H, CH2 pip.), 1.88-1.83 (m Hz, 2H, CH2 pip.). 13C NMR (CDCl3) δ: 156.71 (C=O benzimid.), 137.41 (C-7a), 137.05 (C-4’), 135.62 (C-2), 135.51 (C-1’), 134.24 (C-3 and C-7a benzimid.), 131.45 (C- 3’ and C-5’), 130.88 (C-3” and C-5”), 130.42 (C-1”), 130.06 (C-2” and C-6”), 129.70 (C-2’ and C-6’), 129.48 (C-3a benzimid.), 129.06 (C-4”), 124.06 (C-6 benzimid.), 122.59 (C-5 benzimid.), 122.47 (C-5), 121.77 (C-7 benzimid.), 121.13 (C-4 benzimid.), 116.08 (C- 3a), 112.39 (C-6), 111.31 (C-4), 111.19 (C-7), 64.20 (NCH2), 54.54 (NCH2 pip.), 52.16 (CH pip.), 30.52 (CH2 pip.). MALDI-TOF MS m/z [M+H]+ Calculated for C33H31N4O: 499.250, Found: 499.216. Anal. Calculated for C33H30N4O: C, 79.49; H, 6.06; N, 11.24. Found: C, 79.53; H, 5.97; N, 11.13.
X-ray structure determination
Single crystal X-ray diffraction data for the compound at room temperature was collected on a CAD-4 Enraf-Nonius diffractometer equipped with graphite monochromated CuKα radiation (λ=1.54180 Å). Cell constants and an orientation matrix for data collection were obtained from a least-squares refinement using the setting angles of 25 carefully centered reflections. The cell parameters were then refined using the full set of collected reflections in the range 2.88°<2θ<68.92°. The half sphere of data was collected at 293 K giving rise to 6004 unique reflections, of which 4040 were observed with I/σ(I)>2. The linear absorption coefficient for CuKα is 6.36 cm-1. An empirical absorption correction, based on azimuthal scans of two reflections [18], was applied which resulted in transmission factors ranging from 0.89 to 0.94. The data were corrected for Lorentz and polarization effects. A correction for secondary extinction was applied (coefficient=49 × 10-4). The structure was solved by direct methods and successive Fourier difference syntheses and refined on (F2) by full-matrix least-squares methods respectively with the SHELXS-97 and SHELXL-97 programs [19]. All non-H atoms were refined anisotropically and the positions of the H atoms were deduced from coordinates of the non-H atoms they are linked to, confirmed by Fourier synthesis and treated according to the riding model during refinement. H atoms were included for structure factor calculations, but not refined. The final cycle of full-matrix least-squares refinement was based on 4040 observed reflections [I>2.00σ(I)] and 404 variable parameters and converged with agreement factors of R1=0.064 and wR2=0.180 (All data). All graphical contents have been performed using the OLEX2 interface [20].
The crystal data and refinement details are summarized in Table 1. Crystallographic data (excluding structure factors) for the structural analysis has been deposited with the Cambridge Crystallographic Data Centre, No. CCDC-888295. Copies of this information may be obtained free of charge from: The Director, CCDC, 12 Union Road, Cambridge, CB2 1EZ, UK, Fax: +44(1223)336-033, E-mail: deposit@ccdc.cam.ac.uk or web: www.ccdc.cam.ac.uk.
| Parameter | Value |
|---|---|
| Chemical formula | C33H30N4O, 3(CH3OH) |
| Formula weight | 594.74 |
| T (K) | 293(2) K |
| Wavelength | 1.54180 Å |
| Crystal system, space group | Triclinic, P-1 |
| Unit cell dimensions | a=9.572(7) Å |
| b=11.091(2) Å | |
| c=15.976(4) Å | |
| a= 92.53(2) | |
| β=105.45(3) | |
| g=94.65(3) | |
| Volume | 1625.6(13) Å3 |
| Z, Calculateddensity | 2, 1.215 Mg/m3 |
| Absorption coefficient | 0.636 mm-1 |
| F (000) | 636 |
| Crystal size | 0.18 × 0.15 × 0.10 mm |
| Theta range for data collection | 2.88 to 68.92° |
| Limiting indices | -11<=h<=11, -13<=k<=13, 0<=l<=19 |
| Reflectionscollected/unique | 6004/6004 [R(int)=0.0000] |
| Completeness to theta=68.92 | 99.4% |
| Refinementmethod | Full-matrix least-squares on |
| Data/restraints/parameters | 6004/0/404 |
| Goodness-of-fit on F^2 | 1.019 |
| Final R indices [I>2s(I)] | R1=0.0638, wR2=0.1528 |
| R indices (all data) | R1=0.0973, wR2=0.1800 |
| Largestdiff. peak and hole | 0.351 and -0.279 e.Å-3 |
Table 1: Crystal data, data collection and structure refinement.
Results and Discussion
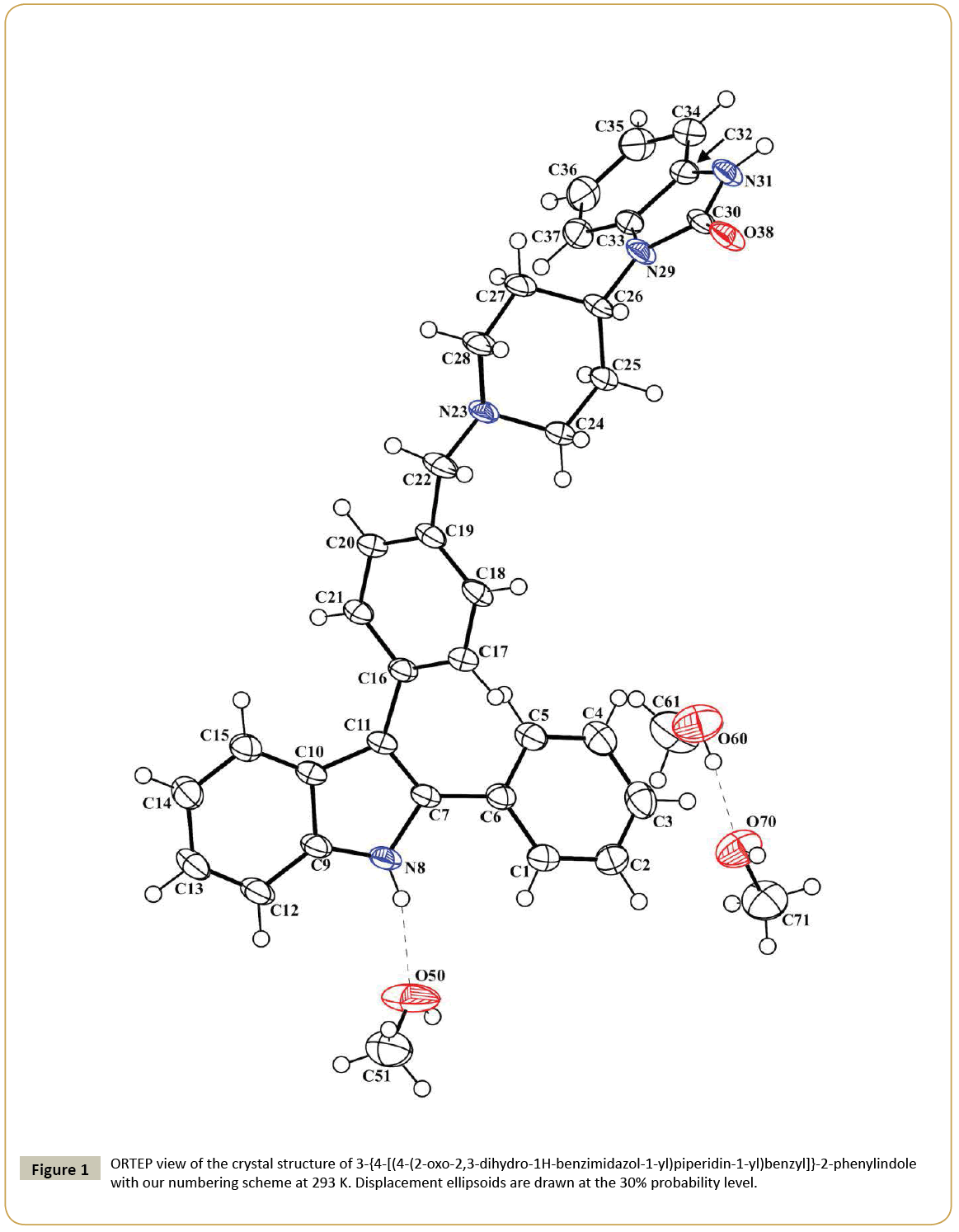
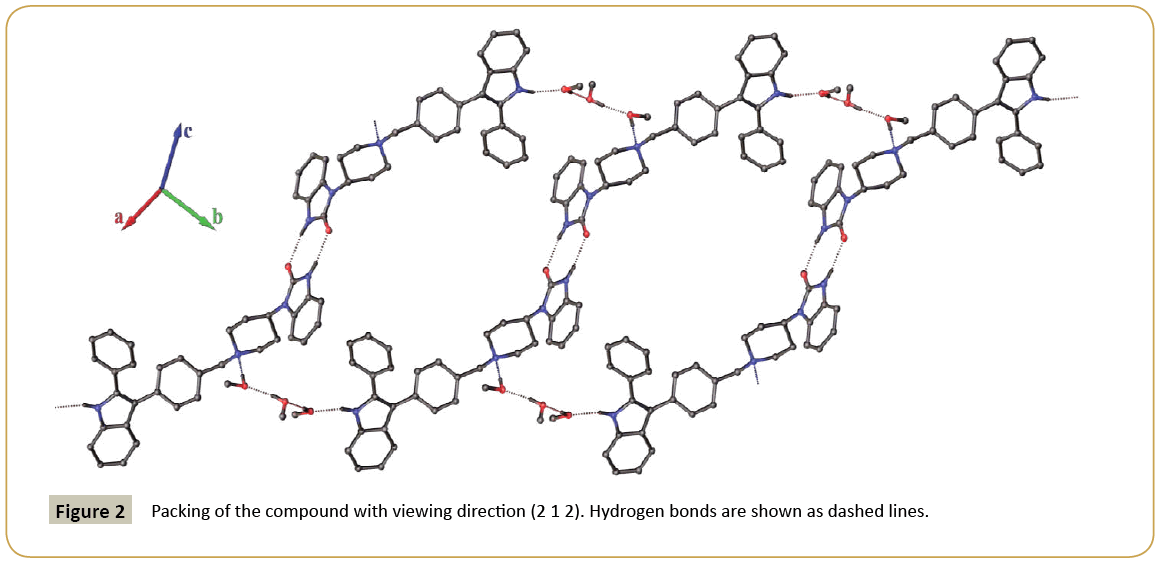
The molecular structure of 3-{4-[(4-(2-oxo-2,3-dihydro-1Hbenzimidazol- 1-yl)piperidin-1-yl)benzyl]}-2-phenylindole is depicted in Figure 1. The 3D spatial determination established by X-ray crystallography confirmed the structure in the solid state as anticipated on the basis of IR, 1H and 13C NMR data. The asymmetric unit of the crystal structure consists in one molecule of the title compound co-crystalized with three methanol molecules. The selected bond distances and torsion angles are collected in Table 2. The indole system is nearly planar; the maximum deviations from the least-squares plane are found for C(10) lying 0.006(2) Å from the benzene ring and for C(11) lying -0.010(2) Å from the pyrrole ring, the dihedral angle between the two ring planes being 0.62(13)°. In the five-membered ring containing atom N(8), the intra-ring bond angles range from 106.5(2)° to 110.0(2)°; the N(8)-C(7) and N(8)-C(9) bond lengths are 1.386(3) and 1.365(3) Å, respectively, which indicates that the geometry around N(8) is normal for sp2 hybridization, as expected for π-conjugation of indole rings previously described [21]. Thus, all atoms that constitute this system have a planar-trigonal configuration, i.e., they are sp2-hybridized. Moreover, the C(10)-C(11) bond, noticed at 1.440(4) Å, is longer than the standard aromatic C-C bond [22] and is in agreement with the values observed in various other indole derivatives [23-25]. The C(1)-C(6) phenyl ring makes a dihedral angle of 32.3(1)° with the mean plane of the indole moiety, and the C(16)-C(21) phenyl group at 3-position makes a dihedral angle of 49.4(1)° with the plane of the indole ring. The torsion angles C(11)-C(7)-C(6)-C(5) and C(7)-C(11)-C(16)-C(17) are ± 31.4(5)° and ± 46.6(4)°, respectively. Presumably, the substituents on C(11) and C(7) in this molecule are turned apart for steric reasons: fairly bulky groups are attached to contiguous carbon atoms of the five-membered ring, in accordance with previously described 2,3-diphenylindole derivatives [25]. The piperidine ring has the chair conformation; the nitrogen atom N(23) and the carbon C(26) are displaced on either side from the least-squares plane C24-C25-C27-C28 by 0.691(4) and 0.656(4) Å, respectively. Moreover, the angles N(23)-C(28)-C(27) and N(23)-C(24)-C(25) are found in the same range 110.8(2) and 111.4(2)°. The 1,3-dihydrobenzimidazol-2-one ring system is almost planar, with the largest deviation being for atom O(38) [0.019(2) Å]. The average bond distances and angles for this 1,3-dihydrobenzimidazol-2-one system are in agreement with those of previous studies on 2-oxo-2,3-dihydro-1H-benzimidazol- 1-yl compounds in the literature [14-17]. Molecules of the title compound are linked by a double H-bonds between the O(38) and N(31) atoms of the imidazole rings to form dimers and H-bonding interactions through the three methanol molecules that connect the dimers together to form chains in the (1 -1 0) direction (Table 3) (Figure 2). The crystal cohesion is ensured by short Van der Waals contacts and C-Hπ interactions in the others directions.
| Bond Distances | Value |
|---|---|
| N(8)-C(7) | 1.386(3) |
| N(8)-C(9) | 1.365(3) |
| C(10)-C(11) | 1.440(4) |
| C(30)-O(38) | 1.229(3) |
| N(31)-C(30) | 1.367(3) |
| N(29)-C(30) | 1.369(3) |
| N(23)-C(24) | 1.467(3) |
| N(23)-C(28) | 1.472(3) |
| N(23)-C(22) | 1.477(3) |
| Bond Angles | Value |
| N(8)-C(7)-C(11) | 108.2(2) |
| C(7)-C(11)-C(10) | 107.5(2) |
| C(11)-C(10)-C(9) | 106.5(2) |
| C(10)-C(9)-N(8) | 107.8(2) |
| C(9)-N(8)-C(7) | 110.0(2) |
| N(23)-C(28)-C(27) | 110.8(2) |
| N(23)-C(24)-C(25) | 111.4(2) |
| Torsion Angles | Value |
| C(11)-C(7)-C(6)-C(5) | ±31.4(5) |
| C(7)-C(11)-C(16)-C(17) | ±46.6(4) |
Table 2: Selected bond distances, angles and torsion angles (Å, ÃÆââ¬Â¹Ãâà ¡) with su’s in parentheses.
| D-HÃÆâÃâââ¬ÂÃâÃâ A | D-H | H-A | D-A | D-H…A |
|---|---|---|---|---|
| N(8)-H(8)ÃÆâÃâââ¬ÂÃâÃâ O(50) | 0.86 | 1.97 | 2.814(4) | 165.1 |
| N(31)-H(31)ÃÆâÃâââ¬ÂÃâÃâ O(38)i | 0.86 | 1.92 | 2.771(3) | 169.4 |
| O(50)-H(50)ÃÆâÃâââ¬ÂÃâÃâ O(60)ii | 0.82 | 1.83 | 2.646(4) | 170.4 |
| O(60)-H(60)ÃÆâÃâââ¬ÂÃâÃâ O(70) | 0.82 | 1.78 | 2.602(4) | 174.2 |
| O(70)-H(70)ÃÆâÃâââ¬ÂÃâÃâ N(23)iii | 0.82 | 1.94 | 2.758(4) | 175.4 |
Table 3: Hydrogen-bonding geometry (Å, °). Symmetry codes: (i) -1-x, 2-y, 1-z, (ii) 1+x, +y, +z, (iii) +x, -1+y, +z.
Conclusion
In the present work, we synthesized the new 3-{4-[(4-(2-oxo- 2,3-dihydro-1H-benzimidazol-1-yl)piperidin-1-yl)benzyl]}- 2-phenylindole which showed antileukemic activity in a submicromolar range on the human leukemic cell lines Jurkat (lymphoid cell line), K562 and HL60 (myeloid cell lines). We report herein on the structural characterization of this new indole scaffold. The present crystal structure determination could now help us to understand the detailed three-dimensional arrangement of this compound, which could be used to generate a broad variety of 3-substituted 2-phenylindoles of interest to medicinal chemists, but will also contribute to the structural database in which there are very few structures containing the indole skeleton. Moreover, solid-state data could be used to clarify the mechanism of action implicating this anti-proliferative derivative.
Acknowledgments
This publication was supported by a Grant from Ligue Nationale contre le Cancer (Comité Aquitaine-Charentes, Bordeaux, France).
References
- Lichtman MA (2008) Battling the hematological malignancies: the 200 year’s war. The Oncologist 13: 126-138.
- Li JJ (2013) Heterocyclic chemistry in drug discovery. John Wiley & Sons, Hoboken.
- Biswal S, Sahoo U, Sethy S, Kumar HKS, Banerjee M (2012) Indole: the molecule of diverse biological activities. Asian J Pharm Clin Res 5: 1-6.
- Shafakat Alia NA, Darab BA, Pradhana V, Farooquia M (2013) Chemistry and Biology of indoles and Indazoles: A mini-review. Mini-Rev Med Chem 13: 1792-1800.
- Sharma V, Kumar V, Pathak D (2010) Biological Importance of the Indole Nucleus in Recent Years: A Comprehensive Review. J Heterocyclic Chem 47: 491-502.
- Kaushik NK, Attri P, Kumar N, Kim CH (2013) Biomedical Importance of Indoles. Molecules 18: 6620-6662.
- Johansson H, Bogelov Jorgensen T, Gloriam DE, Braüner-Osborne H, Sejer Pedersen D (2013) 3-Substituted 2-phenyl-indoles: privileged structures for medicinal chemistry. RSC Adv 3: 945-960.
- Zhang MZ, Chen Q, Yang GF (2015) A review on recent developments of indole-containing antiviral agents. Eur J Med Chem 89: 421-441.
- Sherer C, Snape TJ (2015) Heterocyclic scaffolds as promising anticancer agents against tumours of the central nervous system: Exploring the scope of indole and carbazole derivatives. Eur J Med Chem 97: 552-560.
- Ahmad A, Sakr WA, Rahman KM (2010) Anticancer properties of indole compounds: mechanism of apoptosis induction and role in chemotherapy. Curr Drug Targets 11: 652-666.
- Pelz NF, Bian Z, Zhao B, Shaw S, Tarr JC, et al. (2016) Discovery of 2-Indole-acylsulfonamide Myeloid Cell Leukemia 1 (Mcl-1) Inhibitors Using Fragment-Based Methods. J Med Chem 59: 2054-2066.
- Bai LY, Weng JR, Chiu CF, Wu CY, Yeh SP (2013) OSU-A9, an indole-3-carbinol derivative, induces cytotoxicity in acute myeloid leukemia through reactive oxygen species-mediated apoptosis. Biochem Pharmacol 86: 1430-1440.
- Romagnoli R, Baraldi PG, Carrion MD, Cruz-Lopez O, Cara CL, et al. (2009) Discovery of 8-methoxypyrazino[1,2-a]indole as a New Potent Antiproliferative Agent Against Human Leukemia K562 Cells. A Structure-Activity Relationship Study. Lett Drug Des Discov 6: 298-303.
- Desplat V, Geneste A, Begorre MA, Belisle Fabre S, Brajot S, et al. (2008) Synthesis of New Pyrrolo[1,2-a]quinoxaline Derivatives as Potential Inhibitors of Akt Kinase. J Enzyme Inhib Med Chem 23: 648-658.
- Desplat V, Moreau S, Gay A, Belisle-Fabre S, Thiolat D, et al. (2010) Synthesis and Evaluation of the Antiproliferative Activity of Novel Pyrrolo[1,2-a]quinoxaline Derivatives, Potential Inhibitors of Akt Kinase. Part II. J Enzyme Inhib Med Chem 25: 204-215.
- Desplat V, Moreau S, Belisle-Fabre S, Thiolat D, Uranga J, et al. (2011) Synthesis and Evaluation of the Antiproliferative Activity of Novel Isoindolo[2,1-a]quinoxaline and Indolo[1,2-a]quinoxaline Derivatives. J Enzyme Inhib Med Chem 26: 657-667.
- Desplat V, Vincenzi M, Lucas R, Moreau S, Savrimoutou S, et al. (2016) Synthesis and evaluation of the cytotoxic activity of novel ethyl 4-[4-(4-substitutedpiperidin-1-yl)]benzyl-phenylpyrrolo[1,2-a]quinoxaline-carboxylate derivatives in myeloid and lymphoid leukemia cell lines. Eur J Med Chem 113: 214-227.
- North ACT, Phillips DC, Mathews FS (1968) A semi-empirical method of absorption correction. Acta Crystallogr Sect A 24: 351-359.
- Sheldrick GM (2008) A short history of SHELX. Acta Crystallogr Sect A 64: 112-122.
- Dolomanov OV, Bourhis LJ, Gildea RJ, Howard JAK, Puschmann H (2009) OLEX2: a complete structure solution, refinement and analysis program. J Appl Crystallogr 42: 339-341.
- Huang XH, Zhang QF, Sung HHY (2004) 4-Bromo-N-(2-phenyl-1H-indol-7-yl)-benzenesulfonamide. Acta Crystallogr Sect E 60: o488-489.
- Allen FH, Kennard O, Watson DG, Brammer L, Orpen AG, et al. (1987) Tables of bond lengths determined by X-ray and neutron diffraction. Part 1, Bond lengths in organic compounds. J Chem Soc Perkin Trans II, S1-S19.
- Chandrakantha TN, Puttaraja P, Pattabhi V (1991) Structure of ethyl 5-chloro-1-cyanomethyl-3-phenylindole-2-carboxylate. Acta Crystallogr Sect C 47: 995-998.
- Schmelter B, Bradaczek H, Luger P (1973) Die Kristall- und Molekülstruktur des 2,3-Diphenylindols. Acta Crystallogr Sect B 29: 971-976.
- Phetrak N, Rukkijakan T, Sirijaraensre J, Prabpai S, Kongsaeree P, et al. (2013) Regioselectivity of Larock Heteroannulation: A Contribution from Electronic Properties of Diarylacetylenes. J Org Chem 78: 12703-12709.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences